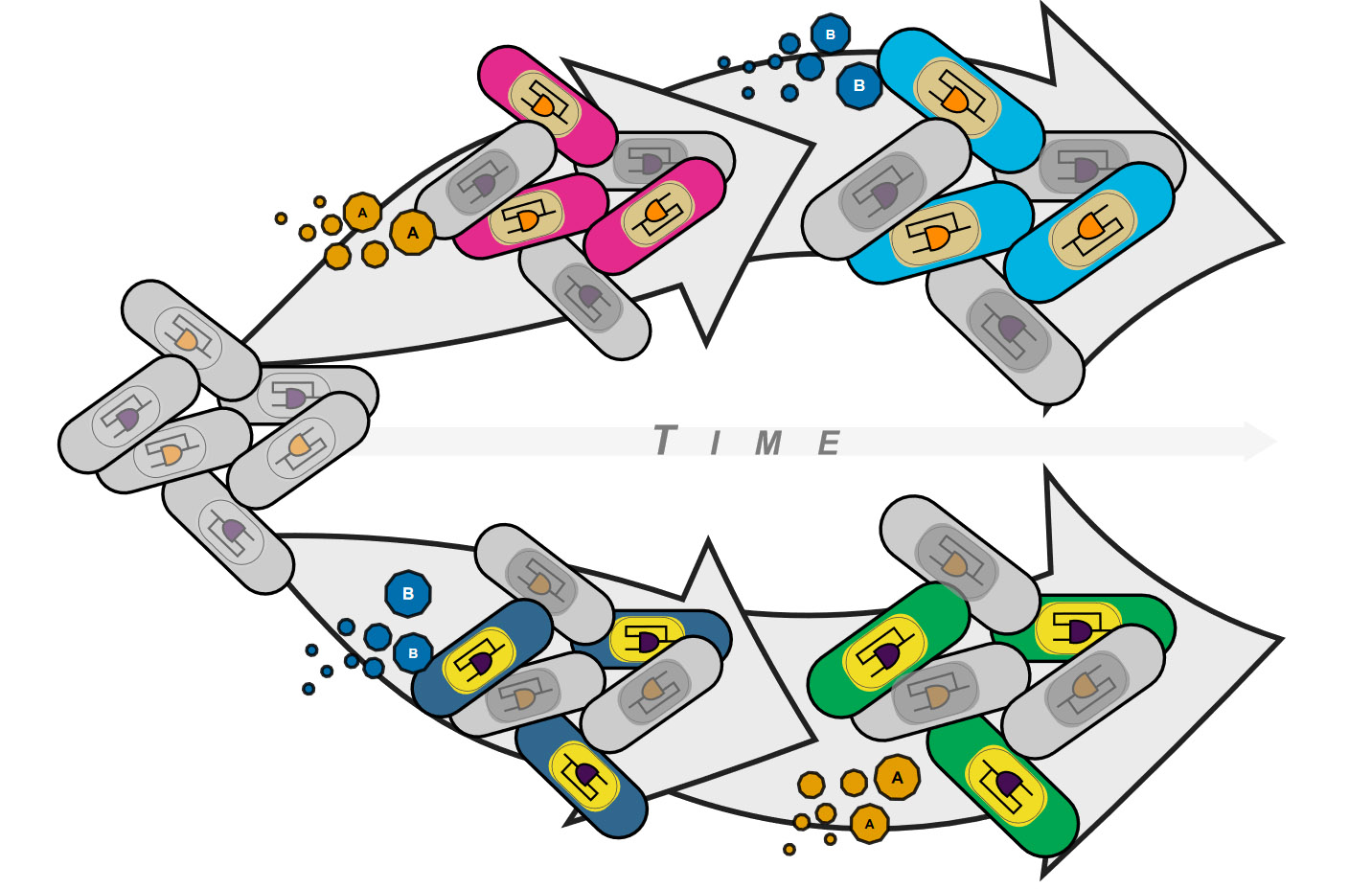
Genetic programs operating in a history-dependent fashion are ubiquitous in nature and govern sophisticated processes such as development and differentiation. The ability to systematically and predictably encode such programs would advance the engineering of synthetic organisms and ecosystems with rich signal processing abilities.
In this paper, researchers from the synthetic biology group at the CBS implemented robust, and scalable history-dependent programs by distributing the computational labor across a cellular population. The group also developed automated workflows that researchers can use to streamline program design and optimization.
These history-dependent cellular programs will support many applications using cellular populations for material engineering, biomanufacturing, and healthcare.
These results have been published in Nature Communications. Together with a 'Behind the paper' blog post shared in Nature Bioengineering Community.
link to the paper:
https://www.nature.com/articles/s41467-020-18455-z
link to the blog post:
https://bioengineeringcommunity.nature.com/posts/history-dependent-logic-in-cellular-populations

